8. Prediction Intervals for Data Streams#
This notebook covers how to utilise prediction intervals for regression tasks in CapyMOA.
Two methods for obtaining prediction intervals are currently available in CapyMOA: MVE and AdaPI.
More details about prediction intervals for streaming data can be found in the AdaPI paper:
More information about CapyMOA can be found at https://www.capymoa.org.
last updated on 28/11/2025
[1]:
from capymoa.datasets import Fried
# load data
fried_stream = Fried()
8.1 Basic prediction interval usage#
Here is an example of prediction intervals in CapyMOA.
Currently available prediction interval learners require a regressor base model.
[2]:
from capymoa.regressor import SOKNL
from capymoa.prediction_interval import MVE
# build prediction interval learner in regular manner
soknl = SOKNL(schema=fried_stream.get_schema(), ensemble_size=10)
mve = MVE(schema=fried_stream.get_schema(), base_learner=soknl)
# build prediction interval learner in in-line manner
mve_inline = MVE(
schema=fried_stream.get_schema(),
base_learner=SOKNL(schema=fried_stream.get_schema(), ensemble_size=10),
)
8.2 Creating evaluators#
There are currently two types of prediction interval evaluators implemented: basic (cumulative) and windowed.
[3]:
from capymoa.evaluation.evaluation import (
PredictionIntervalEvaluator,
PredictionIntervalWindowedEvaluator,
)
# build prediction interval (basic and windowed) evaluators
mve_evaluator = PredictionIntervalEvaluator(schema=fried_stream.get_schema())
mve_windowed_evaluator = PredictionIntervalWindowedEvaluator(
schema=fried_stream.get_schema(), window_size=1000
)
8.3 Running test-then-train/prequential tasks manually#
Don’t forget to train the models (call .train() function) at the end!
[4]:
# run test-then-train/prequential tasks
while fried_stream.has_more_instances():
instance = fried_stream.next_instance()
prediction = mve.predict(instance)
mve_evaluator.update(instance.y_value, prediction)
mve_windowed_evaluator.update(instance.y_value, prediction)
mve.train(instance)
8.4 Results from both evaluators#
[5]:
# show results
print(
f"MVE basic evaluation:\ncoverage: {mve_evaluator.coverage()}, NMPIW: {mve_evaluator.nmpiw()}"
)
print(
f"MVE windowed evaluation in last window:\ncoverage: {mve_windowed_evaluator.coverage()}, \nNMPIW: {mve_windowed_evaluator.nmpiw()}"
)
MVE basic evaluation:
coverage: 97.4, NMPIW: 29.55
MVE windowed evaluation in last window:
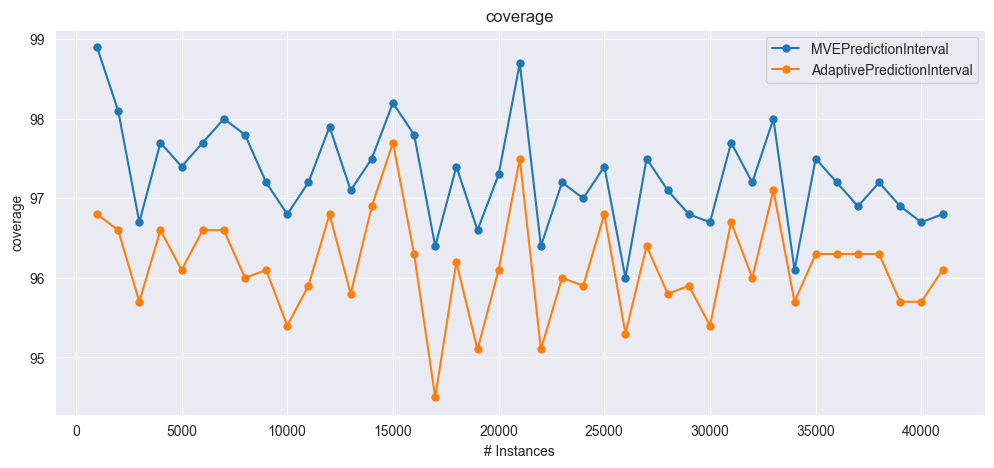
coverage: [99.3, 98.4, 97.0, 98.1, 97.7, 97.3, 98.2, 97.9, 97.7, 96.8, 97.2, 97.5, 97.5, 97.1, 98.2, 97.4, 96.2, 97.2, 97.0, 97.3, 99.0, 96.7, 97.3, 96.9, 97.9, 96.4, 97.9, 97.3, 97.1, 97.1, 97.3, 96.9, 97.1, 97.1, 97.4, 97.3, 97.5, 96.6, 97.3, 97.2],
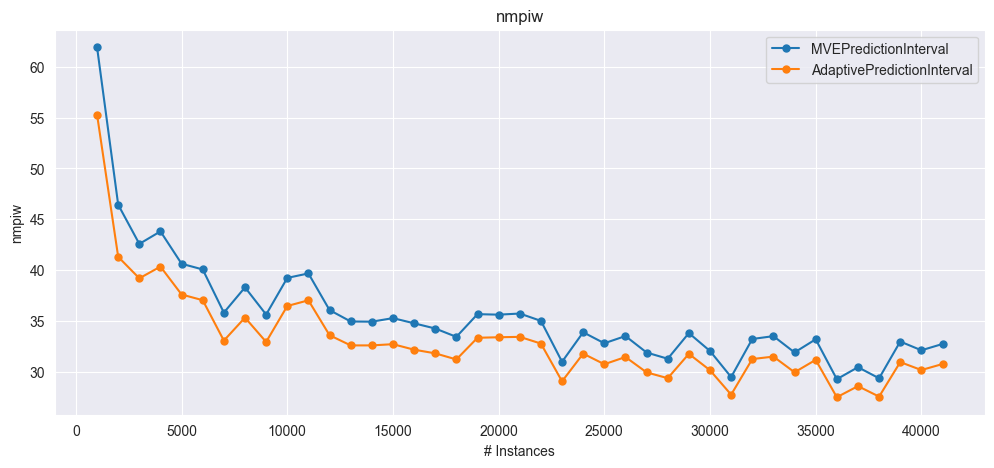
NMPIW: [61.88, 45.51, 41.34, 42.44, 39.27, 38.68, 34.59, 37.0, 34.42, 37.89, 38.35, 34.83, 33.72, 33.68, 34.03, 33.52, 33.01, 32.23, 34.36, 34.27, 34.36, 33.67, 29.79, 32.57, 31.51, 32.19, 30.62, 30.02, 32.44, 30.74, 28.28, 31.85, 32.11, 30.58, 31.8, 28.04, 29.16, 28.12, 31.57, 30.75]
8.5 Wrap things up with prequential evaluation#
Prediction interval tasks also can be wrapped into prequential evaluation in CapyMOA.
[6]:
from capymoa.evaluation import prequential_evaluation
from capymoa.prediction_interval import AdaPI
# restart stream
fried_stream.restart()
# specify regressive model
regressive_learner = SOKNL(schema=fried_stream.get_schema(), ensemble_size=10)
# build prediction interval models
mve_learner = MVE(schema=fried_stream.get_schema(), base_learner=regressive_learner)
adapi_learner = AdaPI(
schema=fried_stream.get_schema(), base_learner=regressive_learner, limit=0.001
)
# gather results
mve_results = prequential_evaluation(
stream=fried_stream, learner=mve_learner, window_size=1000
)
adapi_results = prequential_evaluation(
stream=fried_stream, learner=adapi_learner, window_size=1000
)
# show overall results
print(
f"MVE coverage: {mve_results.cumulative.coverage()}, NMPIW: {mve_results.cumulative.nmpiw()}"
)
print(
f"AdaPI coverage: {adapi_results.cumulative.coverage()}, NMPIW: {adapi_results.cumulative.nmpiw()}"
)
MVE coverage: 97.4, NMPIW: 29.55
AdaPI coverage: 96.25, NMPIW: 27.4
8.6 Plots are also supported#
[7]:
from capymoa.evaluation.visualization import plot_windowed_results
# plot over time comparison
plot_windowed_results(mve_results, adapi_results, metric="coverage")
plot_windowed_results(mve_results, adapi_results, metric="nmpiw")
8.6.1 Plotting prediction intervals over time#
We also provide a visualisation tool for plotting prediction intervals over time.
The function
plot_prediction_intervalcan be used to plot the prediction intervals over time.This function can take one of two prediction interval results as input.
8.6.2 Single result plotting example#
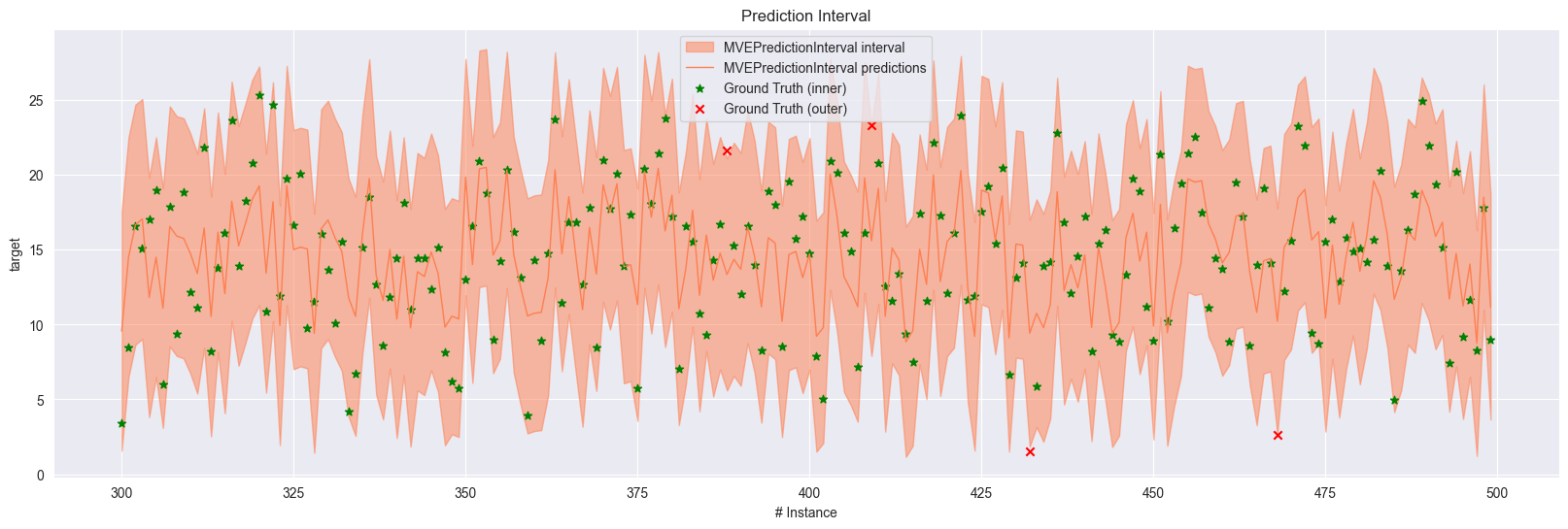
In order to plot the prediction interval over time, we need to have stored the predictions and the ground truth values in the prediction interval results.
The shaded area represents the prediction interval, while the solid line represents the regressor’s predictions.
The star markers represent the ground truth values that are covered by the intervals.
On the other hand, the cross markers represent the ground truth values that are outside the intervals.
The colors can be adjusted by the
colorsparameter in the function as a list.startandendparameters can be used to specify the range of the plot.The
ground truthandpredictionscan be omitted by setting theplot_truthandplot_predictionsparameters toFalse.
We have to set ``optimise`` to ``False`` to avoid subscribing problems.
[8]:
new_mve_learner = MVE(
schema=fried_stream.get_schema(),
base_learner=SOKNL(schema=fried_stream.get_schema(), ensemble_size=10),
)
new_mve_results = prequential_evaluation(
stream=fried_stream,
learner=new_mve_learner,
window_size=1000,
optimise=False,
store_predictions=True,
store_y=True,
)
[9]:
from capymoa.evaluation.visualization import plot_prediction_interval
plot_prediction_interval(new_mve_results, start=300, end=500, colors=["coral"])
8.6.3 Two results plotting example#
For comparison purposes, we can also plot two prediction interval results over time.
We don’t offer the ability to take more than two since it makes the plot too messy to read.
[10]:
# Let's add another results
new_adapi_learner = AdaPI(
schema=fried_stream.get_schema(),
base_learner=SOKNL(schema=fried_stream.get_schema(), ensemble_size=10),
limit=0.001,
)
new_adapi_results = prequential_evaluation(
stream=fried_stream,
learner=new_adapi_learner,
window_size=1000,
optimise=False,
store_predictions=True,
store_y=True,
)
[11]:
plot_prediction_interval(
new_mve_results,
new_adapi_results,
start=300,
end=500,
colors=["coral", "teal"],
plot_predictions=False,
)
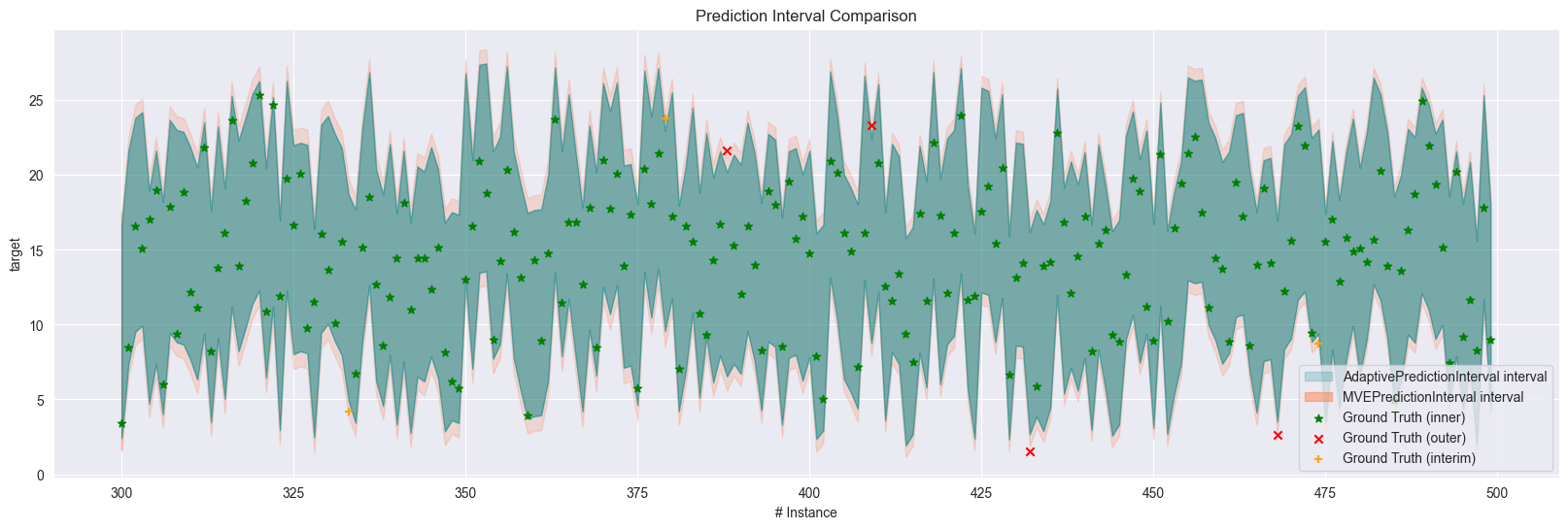
New plus markers represent the ground truth values that are covered by the narrower but not the wider intervals.
The function automatically puts the wider area to the back to make the narrower intervals more visible.